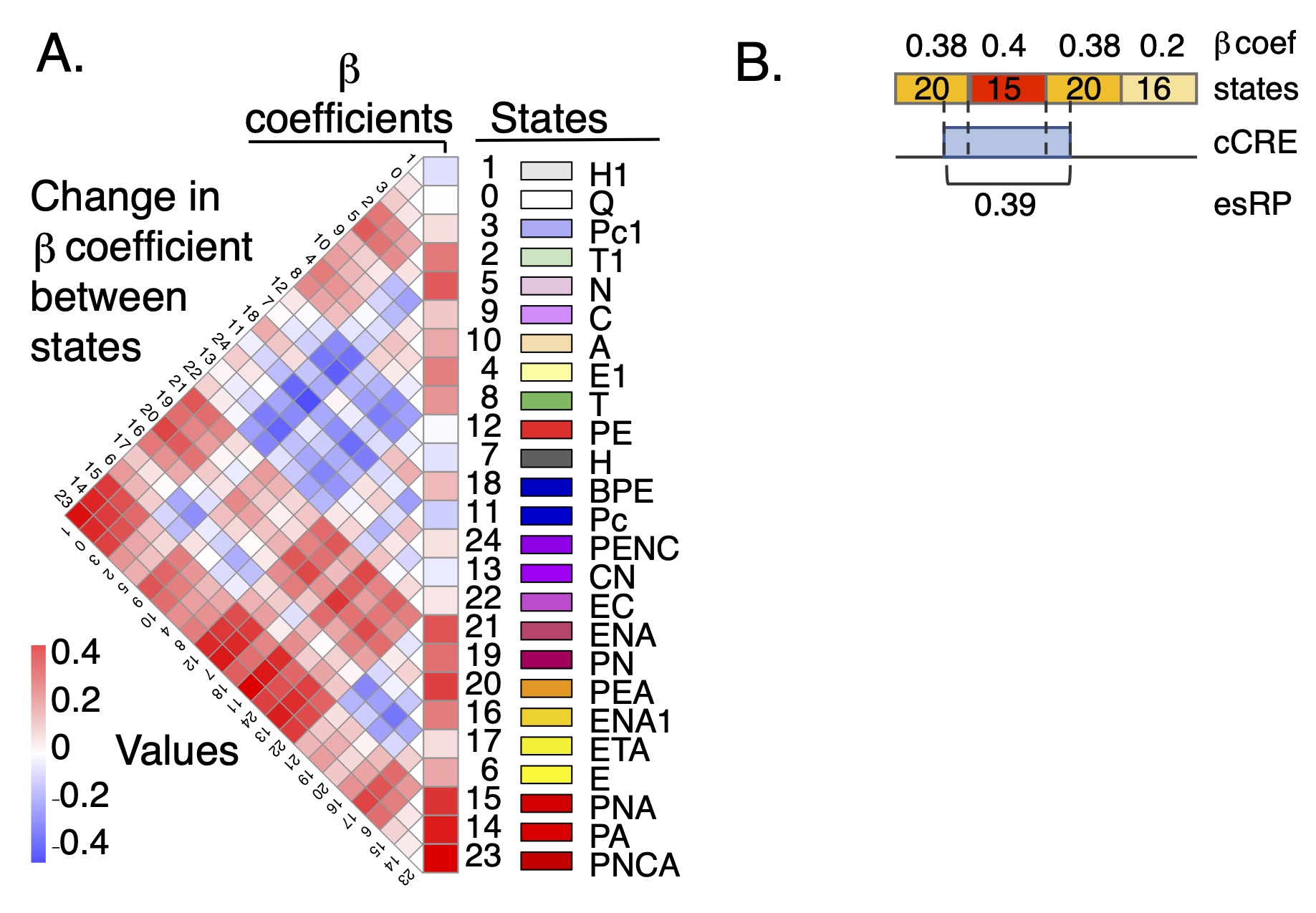
esRP scores for cCREs The epigenetic state Regulatory Potential (esRP) score is an estimate of the contribution of a candidate cis-regulatory element, or cCRE, to the level of expression of a (potential) target gene. The score is derived from the beta coefficients that relate the proportion of cCRE intervals in a particular multi-feature epigenetic state (available in the super track Epigenetic states) to expression levels of target genes in a multivariate linear regression (Xiang et al. 2024; Figure 1). The esRP score is simply a weighted sum of beta coefficients of states that cover a cCRE in a cell type, where the weights are the region covered by different states. The cCREs were defined as DNA intervals with a high signal for chromatin accessibility in any cell type (Xiang et al. 2020 and 2024). They can be visualized using the companion super track Candidate CREs, and they are described more thoroughly on the Track Settings page of that super track.

The most useful setting is "pack" mode, in which the cCREs are labeled with the value of the esRP scores in each cell type. In "dense" mode, the display simply shows the position of each cCRE.
The track names (short name and the end of the long name) give an abbreviation for the blood cell type and the biosamples from the Blueprint Consortium (replicates are from different donors) or the file id after downloading and processing other published data (100xxx). The cell types are HSC = hematopoietic stem cell, MPP = multipotent progenitor cell, LMPP = lymphoid-myeloid primed progenitor cell, CMP = common myeloid progenitor, MEP = megakaryocyte erythrocyte progenitor, GMP = granulocyte monocyte progenitor, CLP = common lymphoid progenitor, CD34_E = erythroblasts generated by in vitro differentiation of CD34+ HSCs, ERY = erythroblast, MK = megakaryocyte, EOS = eosinophil, MONp = primary monocyte, MONc = classical monocyte, NEU = neutrophil, B = B cell, NK = natural killer cell, T_CD4 = CD4+ T cell, T_CD8 = CD8+ T cell, HUDEP = immortalized human umbilical cord blood—derived erythroid progenitor cell lines expressing fetal globin genes (HUDEP1) or adult globin genes (HUDEP2), K562 = a human cancer cell line with some features of early megakaryocytic and erythroid cells. AVE is a track with state assignments based on the average signal for each epigenetic feature across cell types.
The methods for multivariate linear regression of epigenetic states versus expression levels to estimate beta coefficients and the calculation of esRP scores are described on the Track Settings page of the parent super track Epigenetic state Regulatory Potential.
Guanjue Xiang calculated the beta-coefficients and esRP scores. Belinda Giardine generated the track displayed and developed the track hub.
Heuston EF, Keller CA, Lichtenberg J, Giardine B, Anderson SM; NIH Intramural Sequencing Center; Hardison RC, Bodine DM. Establishment of regulatory elements during erythro-megakaryopoiesis identifies hematopoietic lineage-commitment points. Epigenetics Chromatin. 2018 May 28;11(1):22. PMID: 29807547; PMCID: PMC5971425.
Xiang G, Keller CA, Heuston E, Giardine BM, An L, Wixom AQ, Miller A, Cockburn A, Sauria MEG, Weaver K, Lichtenberg J, Göttgens B, Li Q, Bodine D, Mahony S, Taylor J, Blobel GA, Weiss MJ, Cheng Y, Yue F, Hughes J, Higgs DR, Zhang Y, Hardison RC. An integrative view of the regulatory and transcriptional landscapes in mouse hematopoiesis. Genome Res. 2020 Mar;30(3):472-484. PMID: 32132109; PMCID: PMC7111515.
Xiang G, He X, Giardine BM, Isaac KJ, Taylor DJ, McCoy RC, Jansen C, Keller CA, Wixom AQ, Cockburn A, Miller A, Qi Q, He Y, Li Y, Lichtenberg J, Heuston EF, Anderson SM, Luan J, Vermunt MW, Yue F, Sauria MEG, Schatz MC, Taylor J, Göttgens B, Hughes JR, Higgs DR, Weiss MJ, Cheng Y, Blobel GA, Bodine DM, Zhang Y, Li Q, Mahony S, Hardison RC. Interspecies regulatory landscapes and elements revealed by novel joint systematic integration of human and mouse blood cell epigenomes. Genome Res. 2024 Aug 20;34(7):1089-1105. PMID: 38951027; PMCID: PMC11368181.
These data are available for use without restrictions.
Ross Hardison rch8@psu.edu