The VISION Project The VISION project conducted a ValIdated Systematic IntegratiON of epigenetic datasets across progenitor and differentiated blood cell types in mouse and human (Heuston et al. 2018, Xiang et al. 2020, Xiang et al. 2024). The project was carried out by an international group of scientists funded by the National Institute of Diabetes, Digestive, and Kidney Diseases of the National Institutes of Health (grant R24DK106766) and with intramural support from the National Human Genome Research Institute. Key products and results of the project can be visualized on the UCSC Genome Browser using this track hub. The project website provides other servers, databases, and data downloads.
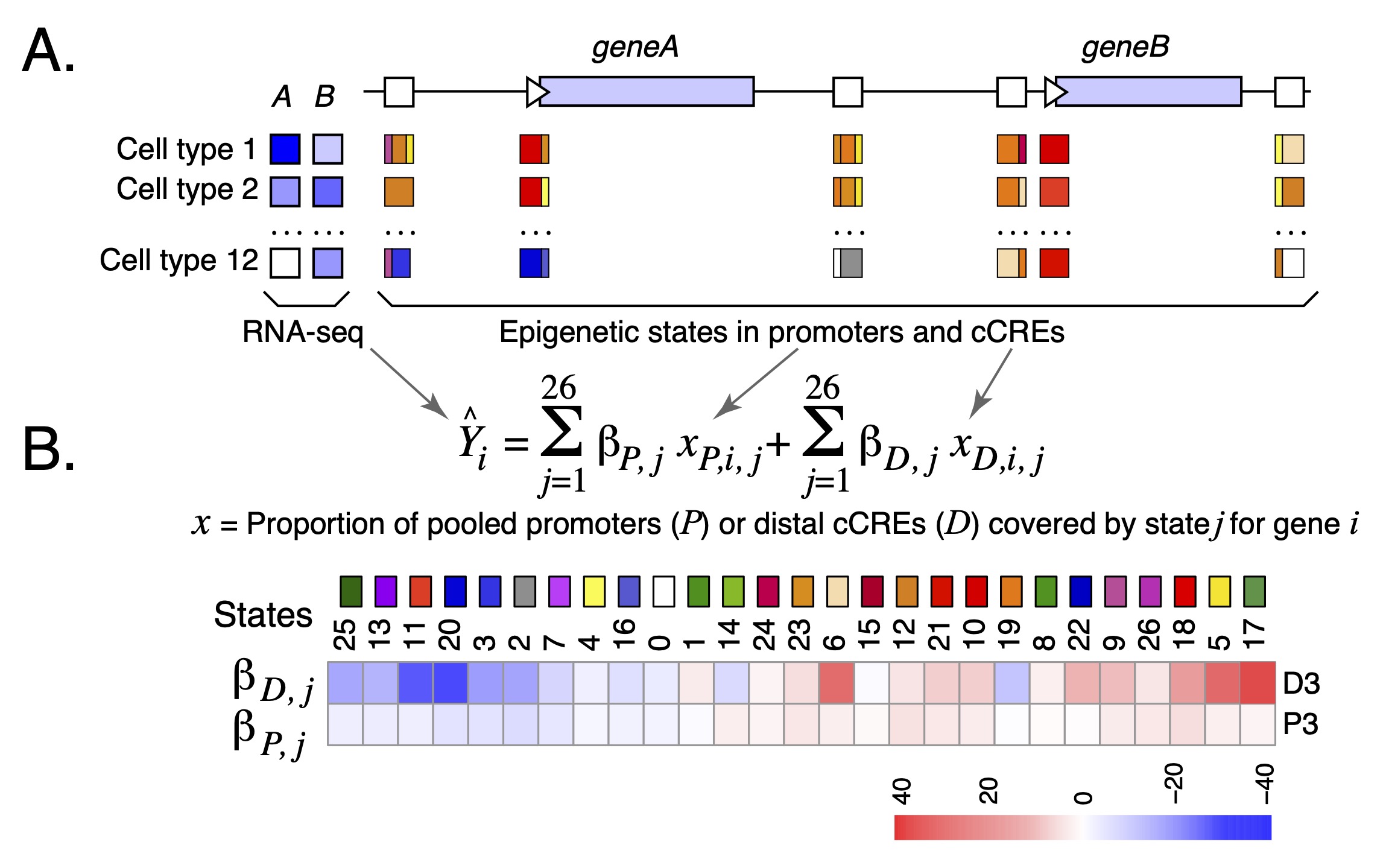
Gene-cCRE pairs Candidate cis-regulatory elements (cCREs) were assigned to potential target genes in a multi-variate regression analysis. The contributions of epigenetic states to variation in expression levels of genes across cell types was estimated by the beta coefficients in the regression analysis (Figure 1). The modeling approach employed in Xiang et al. (2020) included an iterative sub-selection strategy that removed cCREs that had low contributions to explaining expression levels of particular genes. In this way, epigenetic Regulatory Potential (eRP) scores were estimated for each cCRE for each gene in each cell type. The Gene-cCRE pairs composite track provides visualization of the cCREs predicted to potentially regulate a gene and their associated eRP score in each cell type.

The connections between cCREs and the transcription start sites (TSSs) of potential target genes can be visualized in one of three drawing modes: curve, ellipse, or rectangle. The eRP score for that connection can be seen by hovering the computer cursor over the pink circle at the midpoint of the connector. We recommend viewing interactions with both ends in the window as the default for "Show interactions".
The gene-cCRE gene pairs were filtered to remove pairs with no correlation and pairs for which the cCRE covers the TSS for the same gene.
The gene-cCRE pairs are provided for each blood cell type examined. Mouse primary blood cells purified predominantly using cell surface markers include: LSK = Lin-Sca1+Kit+ cells from mouse bone marrow containing hematopoietic stem and progenitor cells, CMP = common myeloid progenitor cell, ERY = erythroblast, GMP = granulocyte monocyte progenitor cell, MONO = monocyte, NEU = neutrophil, CLP = common lymphoid progenitor cell, B = B cell, NK = natural killer cell, T_CD4 = CD4+ T cell, T_CD8 = CD8+ T cell, CFUE = colony forming unit erythroid, fl = designates ERY derived from fetal liver, CFUMK = colony forming unit megakaryocyte, MK imm = immature megakaryocyte from adult bone marrow, MK mat = maturing megakaryocyte derived from fetal liver.
Data from several immortalized cell lines were included. The G1E cells are an immortalized, GATA1-null cell line derived from mouse embryonic stem cells by gene targeting; these cells proliferate in culture as immature erythroid progenitor cells (Weiss, Yu, Orkin 1997). A stable subline of these cells, called G1E-ER4, undergoes terminal erythroid maturation when GATA1 function is restored as an activatable fusion of GATA1 to the ligand-binding domain of the estrogen receptor (ER). Untreated G1E-ER4 cells, carrying the inactive GATA1-ER, proliferate without differentiation, but treatment with estradiol (E2) activates the hybrid protein, effectively complementing the GATA1 loss-of-function and allowing synchronous erythroid differentiation and maturation (Gregory et al. 1999). HPC7 cells are an immortalized line that serves as a model for mouse hematopoietic progenitor cells (Pinto do O 2002). These cells are capable of differentiation in vitro into more mature myeloid cells.
The procedures and modeling for estimating the gene-cCRE pairs are described in Xiang et al. (2020) and its Supplemental Information.
Yu Zhang developed the strategy and conducted the iterative sub-selection approach. Guanjue Xiang, Michael Sauria, Kathryn Isaac, and Alexander Wixom refined the approach and developed the gene-cCRE pairs. Belinda Giardine generated the track and developed the track hub.
Gregory T, Yu C, Ma A, Orkin SH, Blobel GA, Weiss MJ. GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating bcl-xL expression. Blood. 1999; 94:87-96. PMID: 10381501.
Heuston EF, Keller CA, Lichtenberg J, Giardine B, Anderson SM; NIH Intramural Sequencing Center; Hardison RC, Bodine DM. Establishment of regulatory elements during erythro-megakaryopoiesis identifies hematopoietic lineage-commitment points. Epigenetics Chromatin. 2018 May 28;11(1):22. PMID: 29807547; PMCID: PMC5971425.
Pinto do O P, Richter K, Carlsson L. Hematopoietic progenitor/stem cells immortalized by Lhx2 generate functional hematopoietic cells in vivo. Blood. 2002 Jun 1;99(11):3939-46. doi: 10.1182/blood.v99.11.3939. PMID: 12010792.
Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997; 17:1642-1651. PMID: 9032291; PMCID: PMC231889.
Xiang G, Keller CA, Heuston E, Giardine BM, An L, Wixom AQ, Miller A, Cockburn A, Sauria MEG, Weaver K, Lichtenberg J, Göttgens B, Li Q, Bodine D, Mahony S, Taylor J, Blobel GA, Weiss MJ, Cheng Y, Yue F, Hughes J, Higgs DR, Zhang Y, Hardison RC. An integrative view of the regulatory and transcriptional landscapes in mouse hematopoiesis. Genome Res. 2020 Mar;30(3):472-484. PMID: 32132109; PMCID: PMC7111515.
Xiang G, He X, Giardine BM, Isaac KJ, Taylor DJ, McCoy RC, Jansen C, Keller CA, Wixom AQ, Cockburn A, Miller A, Qi Q, He Y, Li Y, Lichtenberg J, Heuston EF, Anderson SM, Luan J, Vermunt MW, Yue F, Sauria MEG, Schatz MC, Taylor J, Göttgens B, Hughes JR, Higgs DR, Weiss MJ, Cheng Y, Blobel GA, Bodine DM, Zhang Y, Li Q, Mahony S, Hardison RC. Interspecies regulatory landscapes and elements revealed by novel joint systematic integration of human and mouse blood cell epigenomes. Genome Res. 2024 Aug 20;34(7):1089-1105. PMID: 38951027; PMCID: PMC11368181.
These data are available for use without restrictions.
Ross Hardison rch8@psu.edu