Differential expression computed with EBSeq along branches and pairwise. These tracks show the differential expression of genes in an ordered set of RNA-seq analyses along several branches of myeloid hematopoietic differentiation in mouse. The total RNA-seq data were used as input to compute differential expression using EBSeq-HMM (Leng et al. 2015).
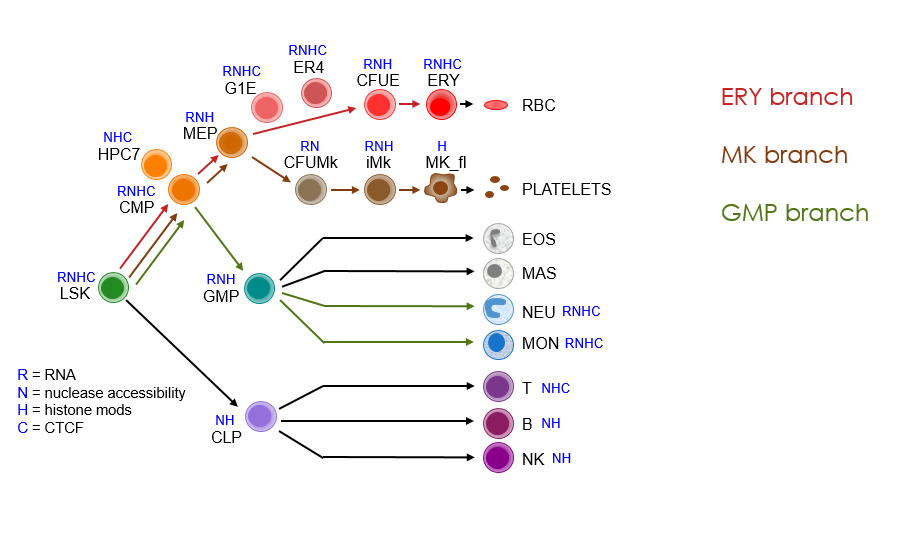
The branches and sub-branches analyzed are indicated in the track names. They can be visualized in the following figure.

The expression levels for differentially expressed genes are visualized as bar plots aligned with each gene model in the browser. Hovering the computer mouse over each bar will give the expression value in each cell type. Clicking on the image on the browser track takes you to a page with a full bar plot for expression levels for each gene in each cell type. Raw and normalized FPKMs can also be visualized.
Mouse primary blood cells purified predominantly using cell surface markers include: LSK = Lin-Sca1+Kit+ cells from mouse bone marrow containing hematopoietic stem and progenitor cells, CMP = common myeloid progenitor cell, MEP = megakaryocyte-erythrocyte progenitor cell, ERY = erythroblast, GMP = granulocyte monocyte progenitor cell, MON = monocyte, NEU = neutrophil, CFUE = colony forming unit erythroid, CFUMK = colony forming unit megakaryocyte, iMK = immature megakaryocyte, MK_fl = megakaryocyte derived from fetal liver.
The G1E cells are an immortalized, GATA1-null cell line derived from mouse embryonic stem cells by gene targeting; these cells proliferate in culture as immature erythroid progenitor cells (Weiss, Yu, Orkin 1997). A stable subline of these cells, called G1E-ER4, undergoes terminal erythroid maturation when GATA1 function is restored as an activatable fusion of GATA1 to the ligand-binding domain of the estrogen receptor (ER). Untreated G1E-ER4 cells, carrying the inactive GATA1-ER, proliferate without differentiation, but treatment with estradiol (E2) activates the hybrid protein, effectively complementing the GATA1 loss-of-function and allowing synchronous erythroid differentiation and maturation (Gregory et al. 1999).
The computer tool EBSeq-HMM (Leng et al. 2015) was used to analyze differential expression for genes in cell types ordered by their position in the classic hierarchical tree of hematopoietic differentiation.
Belinda Giardine ran the differential expression analysis, generated the tracks displayed, and developed the track hub.
Gregory T, Yu C, Ma A, Orkin SH, Blobel GA, Weiss MJ. GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating bcl-xL expression. Blood. 1999; 94:87-96. PMID: 10381501.
Leng N, Li Y, McIntosh BE, Nguyen BK, Duffin B, Tian S, Thomson JA, Dewey CN, Stewart R, Kendziorski C. EBSeq-HMM: a Bayesian approach for identifying gene-expression changes in ordered RNA-seq experiments. Bioinformatics. 2015 Aug15;31(16):2614-22. doi: 10.1093/bioinformatics/btv193. Epub 2015 Apr 5. PMID: 25847007; PMCID: PMC4528625.
Weiss MJ, Yu C, Orkin SH. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol Cell Biol. 1997; 17:1642-1651. PMID: 9032291; PMCID: PMC231889.
Xiang G, Keller CA, Heuston E, Giardine BM, An L, Wixom AQ, Miller A, Cockburn A, Sauria MEG, Weaver K, Lichtenberg J, Göttgens B, Li Q, Bodine D, Mahony S, Taylor J, Blobel GA, Weiss MJ, Cheng Y, Yue F, Hughes J, Higgs DR, Zhang Y, Hardison RC. An integrative view of the regulatory and transcriptional landscapes in mouse hematopoiesis. Genome Res. 2020 Mar;30(3):472-484. PMID: 32132109; PMCID: PMC7111515.
These data are available for use without restrictions.
Ross Hardison rch8@psu.edu